6. CHAPTER 6. THE LEAST SQUARE ESTIMATOR#
SET UP
library(foreign) # to open stata.dta files
library(psych) # for better sammary of descriptive statistics
library(repr) # to combine graphs with adjustable plot dimensions
options(repr.plot.width = 12, repr.plot.height = 6) # Plot dimensions (in inches)
options(width = 150) # To increase character width of printed output
6.1. 6.2 EXAMPLES OF SAMPLING FROM A POPULATION#
rm(list=ls())
df = read.dta(file = "Dataset/AED_GENERATEDDATA.DTA")
attach(df)
df_desc<-describe(df)
print(df_desc)
vars n mean sd median trimmed mad min max range skew kurtosis se
x 1 5 3.00 1.58 3.00 3.00 1.48 1.00 5.00 4.00 0.00 -1.91 0.71
Eygivenx 2 5 7.00 3.16 7.00 7.00 2.97 3.00 11.00 8.00 0.00 -1.91 1.41
u 3 5 -1.03 1.75 -1.63 -1.03 1.29 -2.51 1.69 4.20 0.54 -1.66 0.78
y 4 5 5.97 1.90 4.69 5.97 0.29 4.49 8.61 4.12 0.40 -2.03 0.85
6.1.1. Table 6.1#
print(df.head<-head(df))
x Eygivenx u y
1 1 3 1.6898891 4.689889
2 2 5 -0.3187171 4.681283
3 3 7 -2.5066674 4.493333
4 4 9 -1.6332803 7.366720
5 5 11 -2.3907640 8.609236
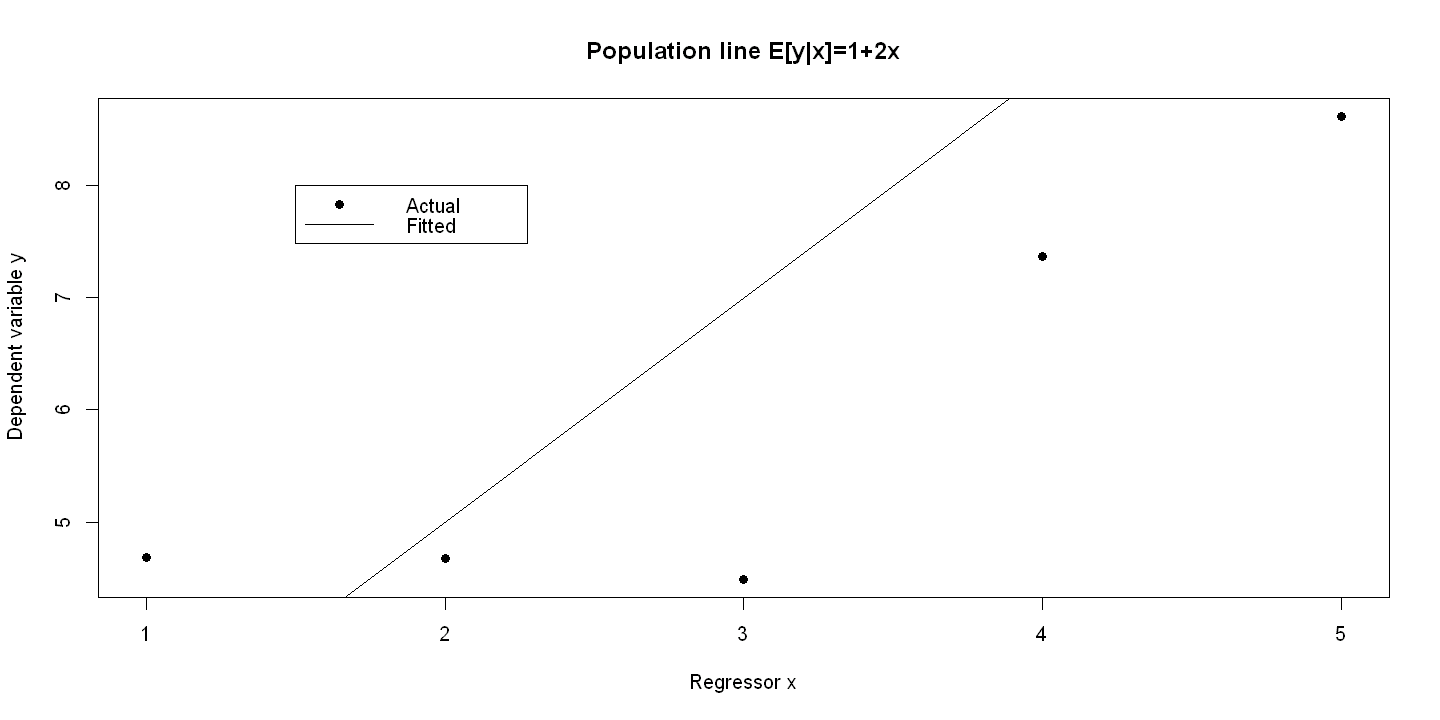
6.1.2. Figure 6.2: panel A#
summary(olsEyx <- lm(Eygivenx ~ x))
plot(x, y, xlab="Regressor x", ylab="Dependent variable y", pch=19, main="Population line E[y|x]=1+2x")
abline(olsEyx)
legend(1.5, 8, c("Actual", "Fitted"), lty=c(-1,1), pch=c(19,-1), bty="o")
Warning message in summary.lm(olsEyx <- lm(Eygivenx ~ x)):
"essentially perfect fit: summary may be unreliable"
Call:
lm(formula = Eygivenx ~ x)
Residuals:
1 2 3 4 5
7.134e-19 6.054e-16 -7.714e-16 -2.760e-16 4.414e-16
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.000e+00 6.723e-16 1.487e+15 <2e-16 ***
x 2.000e+00 2.027e-16 9.867e+15 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.41e-16 on 3 degrees of freedom
Multiple R-squared: 1, Adjusted R-squared: 1
F-statistic: 9.736e+31 on 1 and 3 DF, p-value: < 2.2e-16
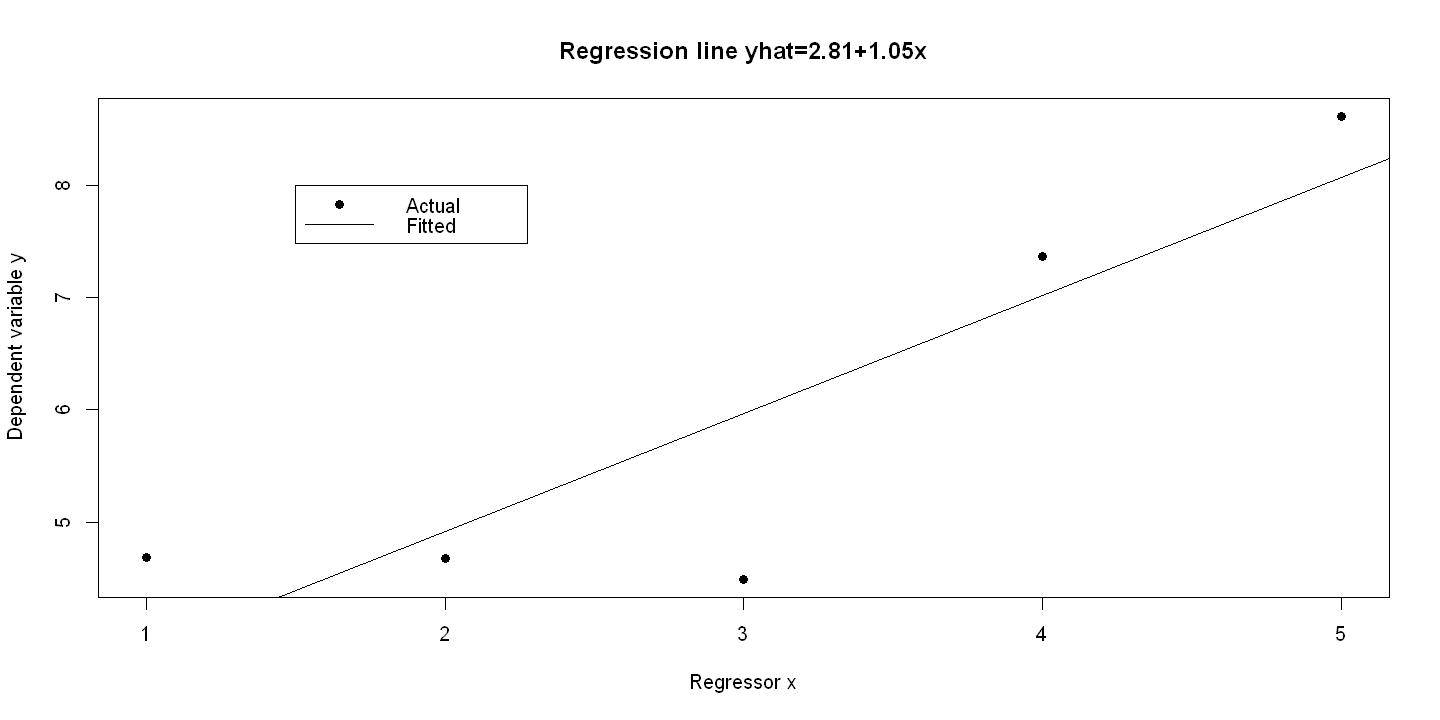
6.1.3. Figure 6.2: panel B#
olsyx <- lm(y ~ x)
summary(olsyx)
plot(x, y, xlab="Regressor x", ylab="Dependent variable y",pch=19, main = "Regression line yhat=2.81+1.05x" )
abline(olsyx)
legend(1.5, 8, c("Actual", "Fitted"), lty=c(-1,1), pch=c(19,-1), bty="o")
Call:
lm(formula = y ~ x)
Residuals:
1 2 3 4 5
0.8266 -0.2344 -1.4748 0.3462 0.5363
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.8109 1.1034 2.547 0.0841 .
x 1.0524 0.3327 3.163 0.0507 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.052 on 3 degrees of freedom
Multiple R-squared: 0.7693, Adjusted R-squared: 0.6925
F-statistic: 10.01 on 1 and 3 DF, p-value: 0.05074
Three regressions from the same model
rm(list=ls())
set.seed(12345)
u = runif(30,min=0,max=1)
x1 = rnorm(30,3,1)
u1 = rnorm(30,0,2)
y1 = 1 + 2*x1 + u1
x2 = rnorm(30,3,1)
u2 = rnorm(30,0,2)
y2 = 1 + 2*x2 + u2
x3 = rnorm(30,3,1)
u3 = rnorm(30,0,2)
y3 = 1 + 2*x3 + u3
ols1 <- lm(y1 ~ x1)
summary(ols1)
ols2 <- lm(y2 ~ x2)
summary(ols2)
ols3 <- lm(y3 ~ x3)
summary(ols3)
Call:
lm(formula = y1 ~ x1)
Residuals:
Min 1Q Median 3Q Max
-4.783 -1.612 -0.090 1.693 4.003
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.3789 1.3276 -0.285 0.777
x1 2.5173 0.3806 6.614 3.56e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.442 on 28 degrees of freedom
Multiple R-squared: 0.6097, Adjusted R-squared: 0.5958
F-statistic: 43.75 on 1 and 28 DF, p-value: 3.565e-07
Call:
lm(formula = y2 ~ x2)
Residuals:
Min 1Q Median 3Q Max
-4.623 -1.528 -0.207 2.122 4.896
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.2469 1.5805 0.789 0.436775
x2 2.0538 0.4584 4.480 0.000115 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.528 on 28 degrees of freedom
Multiple R-squared: 0.4176, Adjusted R-squared: 0.3968
F-statistic: 20.07 on 1 and 28 DF, p-value: 0.0001146
Call:
lm(formula = y3 ~ x3)
Residuals:
Min 1Q Median 3Q Max
-3.9398 -0.9904 0.1098 1.0261 3.8738
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.7696 1.2548 2.207 0.03567 *
x3 1.5345 0.4433 3.461 0.00174 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.927 on 28 degrees of freedom
Multiple R-squared: 0.2997, Adjusted R-squared: 0.2747
F-statistic: 11.98 on 1 and 28 DF, p-value: 0.001742
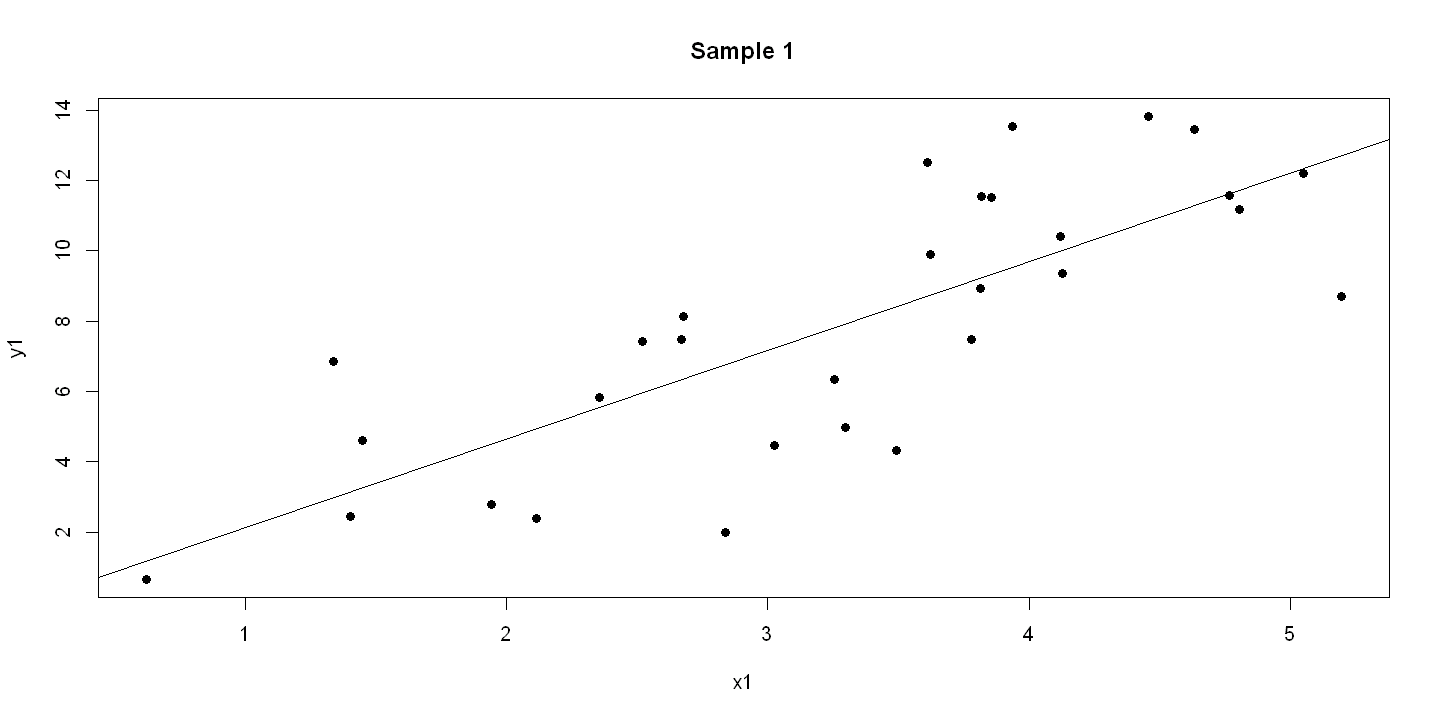
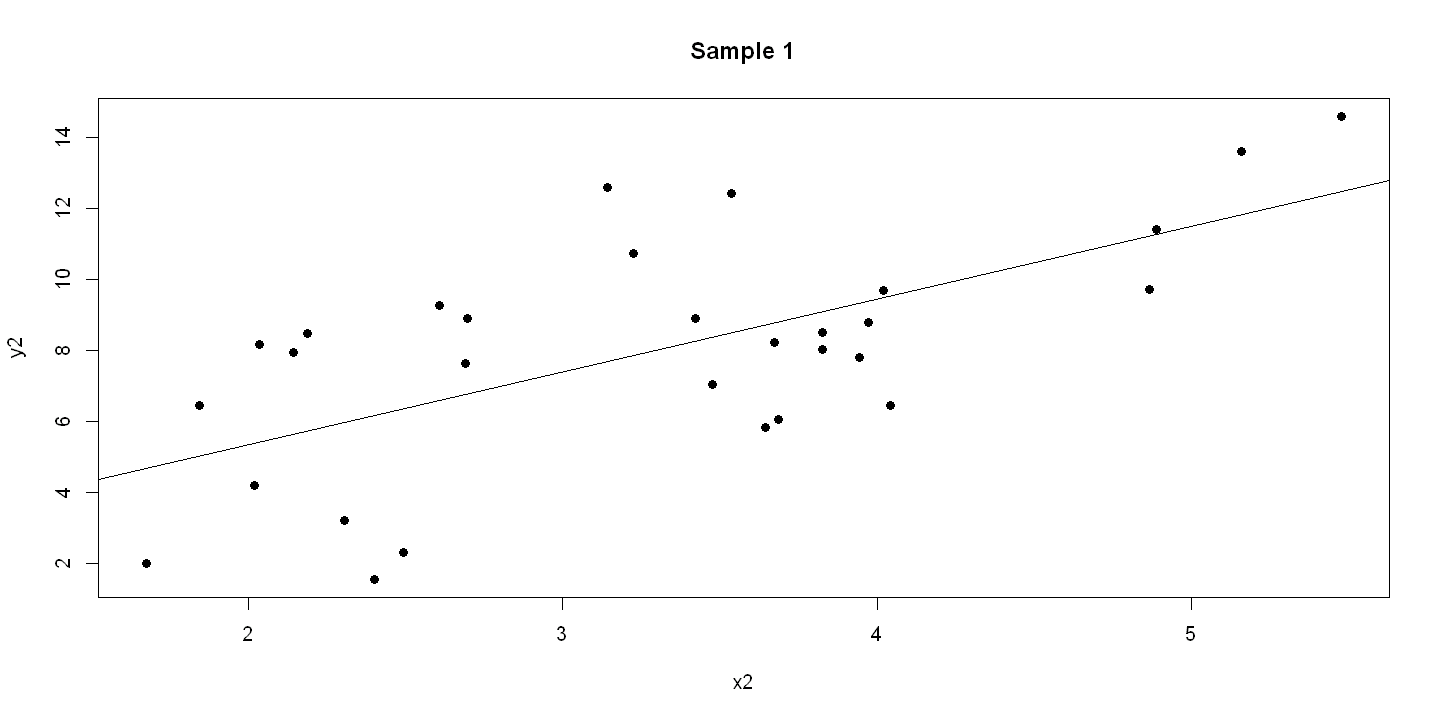
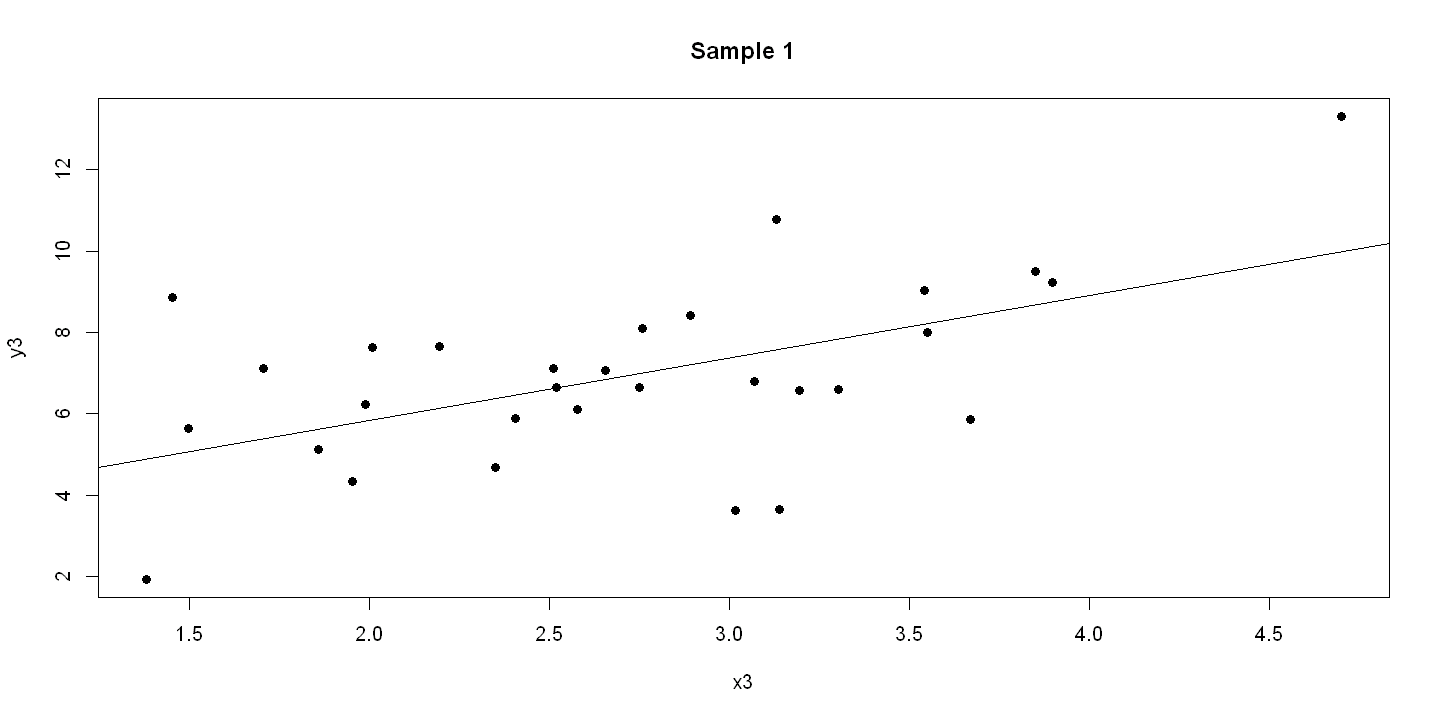
6.1.4. Figure 6.3 - three panels#
plot(x1, y1, pch=19, main = "Sample 1" )
abline(ols1)
plot(x2, y2, pch=19, main = "Sample 1" )
abline(ols2)
plot(x3, y3, pch=19, main = "Sample 1" )
abline(ols3)
Generated data: results of regression on 400 samples of size 30
df = read.dta(file = "Dataset/AED_GENERATEDREGRESSIONS.DTA")
attach(df)
df_desc<-describe(df)
print(df_desc)
head(df)
vars n mean sd median trimmed mad min max range skew kurtosis se
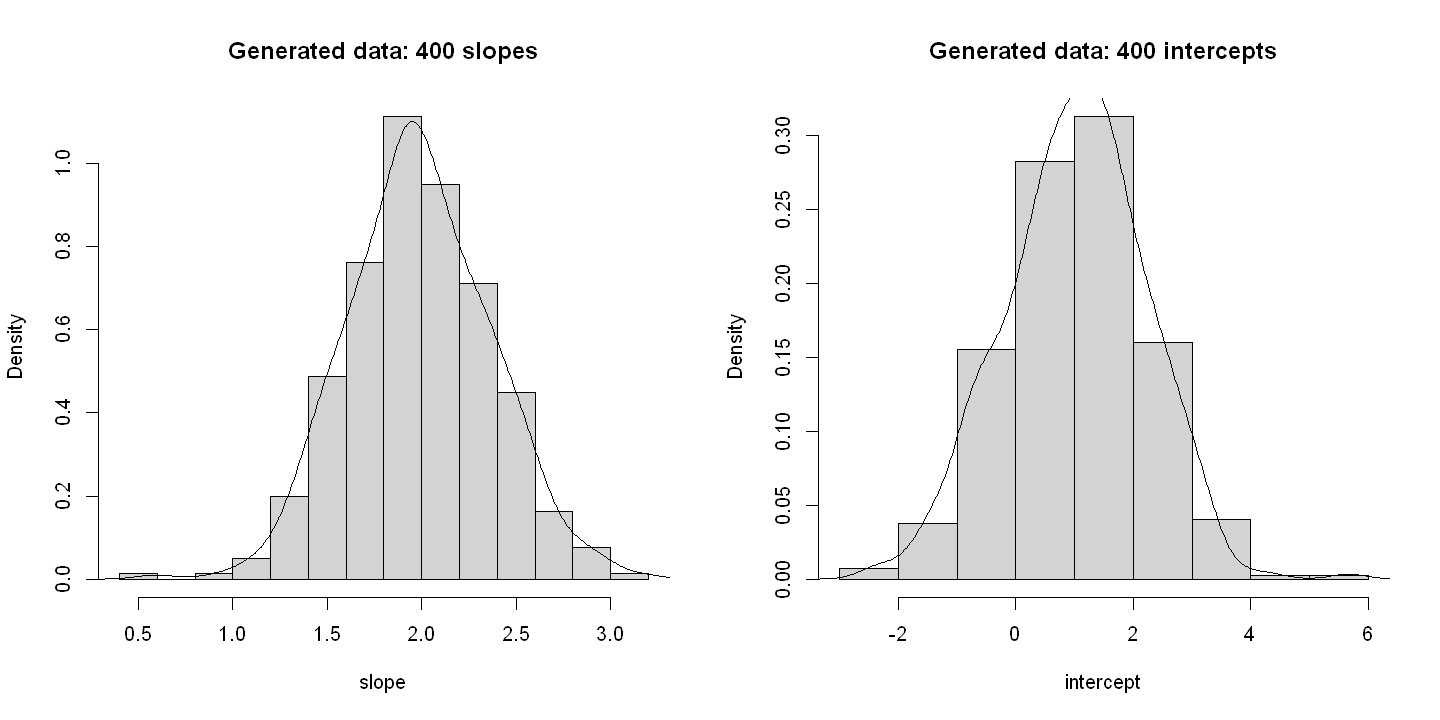
slope 1 400 1.99 0.38 1.96 1.98 0.38 0.59 3.17 2.58 0.04 0.19 0.02
seslope 2 400 0.38 0.07 0.37 0.37 0.07 0.19 0.65 0.46 0.60 0.42 0.00
intercept 3 400 1.02 1.19 1.06 1.03 1.13 -2.46 5.67 8.13 -0.05 0.16 0.06
seintercept 4 400 1.19 0.23 1.17 1.18 0.22 0.63 2.00 1.37 0.60 0.47 0.01
rsquared 5 400 0.50 0.12 0.51 0.50 0.12 0.09 0.82 0.73 -0.40 0.17 0.01
numobs 6 400 30.00 0.00 30.00 30.00 0.00 30.00 30.00 0.00 NaN NaN 0.00
| slope | seslope | intercept | seintercept | rsquared | numobs | |
|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 2.011422 | 0.2921605 | 0.9837164 | 0.9583587 | 0.6286392 | 30 |
| 2 | 1.634170 | 0.3506421 | 1.7954644 | 1.1950867 | 0.4368498 | 30 |
| 3 | 2.094961 | 0.4156610 | -0.0209372 | 1.3890935 | 0.4756781 | 30 |
| 4 | 1.972498 | 0.2285200 | 0.6164197 | 0.8136204 | 0.7268423 | 30 |
| 5 | 2.548882 | 0.6503775 | -0.6771045 | 1.9628325 | 0.3542315 | 30 |
| 6 | 2.064110 | 0.3353717 | 1.2542309 | 0.9950362 | 0.5749864 | 30 |
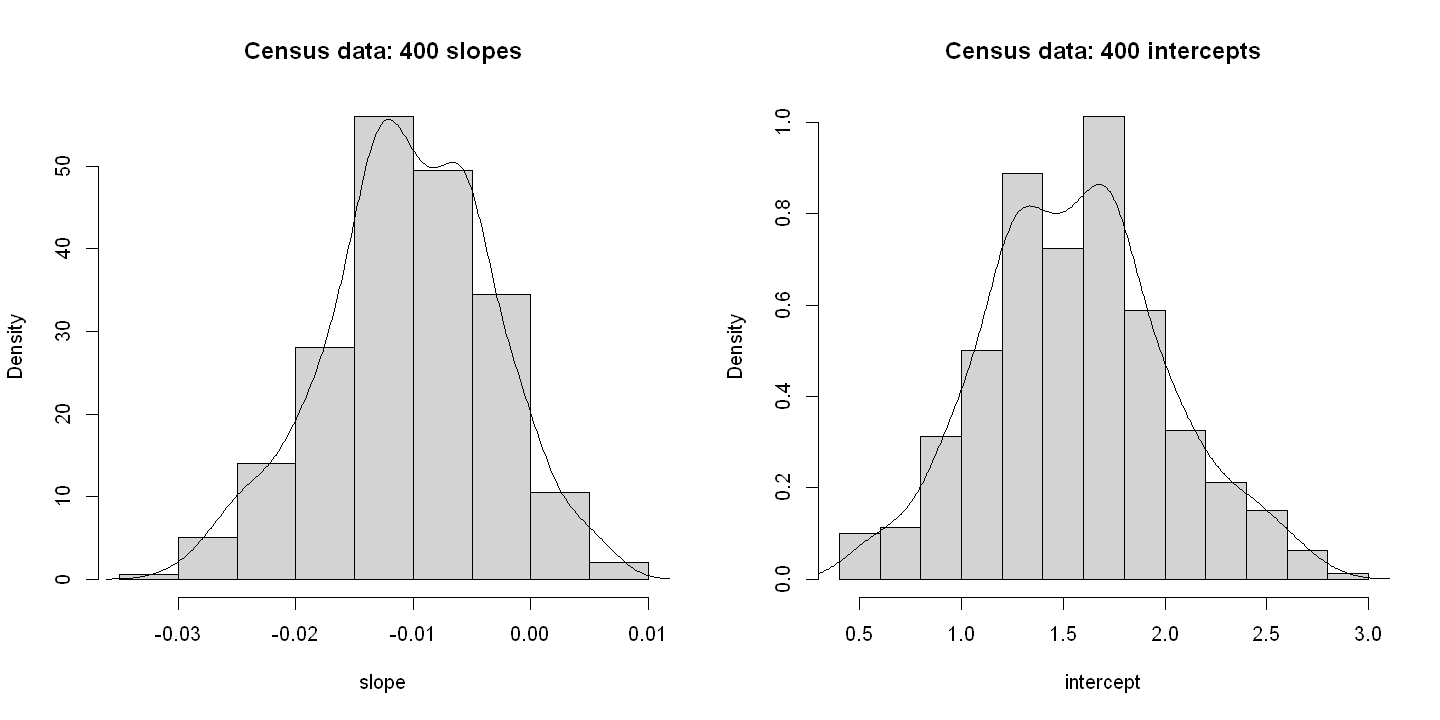
6.1.5. Figure 6.4#
par(mfrow=c(1,2))
# Panel A
hist(slope, freq=FALSE, xlab="slope", main="Generated data: 400 slopes")
lines(density(slope))
# Panel B
hist(intercept, prob=TRUE, xlab="intercept", main="Generated data: 400 intercepts")
lines(density(intercept))
par(mfrow = c(1,1))
Census data: results of regression on 400 samples of size 120
df = read.dta(file = "Dataset/AED_CENSUSREGRESSIONS.DTA")
attach(df)
df_desc<-describe(df)
print(df_desc)
head(df)
The following objects are masked from df (pos = 3):
intercept, numobs, seintercept, seslope, slope
vars n mean sd median trimmed mad min max range skew kurtosis se
slope 1 400 -0.01 0.01 -0.01 -0.01 0.01 -0.03 0.01 0.04 -0.18 -0.15 0.00
seslope 2 400 0.01 0.00 0.01 0.01 0.00 0.00 0.01 0.00 -0.11 -0.02 0.00
intercept 3 400 1.56 0.45 1.55 1.55 0.43 0.45 2.80 2.36 0.17 -0.14 0.02
seintercept 4 400 0.41 0.04 0.42 0.41 0.04 0.31 0.53 0.23 -0.12 -0.03 0.00
numobs 5 400 200.00 0.00 200.00 200.00 0.00 200.00 200.00 0.00 NaN NaN 0.00
| slope | seslope | intercept | seintercept | numobs | |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | -0.01326593 | 0.007451856 | 1.707933 | 0.4797598 | 200 |
| 2 | -0.01021786 | 0.006931265 | 1.534607 | 0.4446451 | 200 |
| 3 | -0.00404300 | 0.006063364 | 1.174318 | 0.3894077 | 200 |
| 4 | -0.01864705 | 0.005981046 | 2.090928 | 0.3841836 | 200 |
| 5 | -0.01285031 | 0.006403644 | 1.704155 | 0.4113336 | 200 |
| 6 | -0.01622651 | 0.006049114 | 1.949338 | 0.3898587 | 200 |